A quantitative framework for evaluating data structure preservation by dimensionality reduction techniques
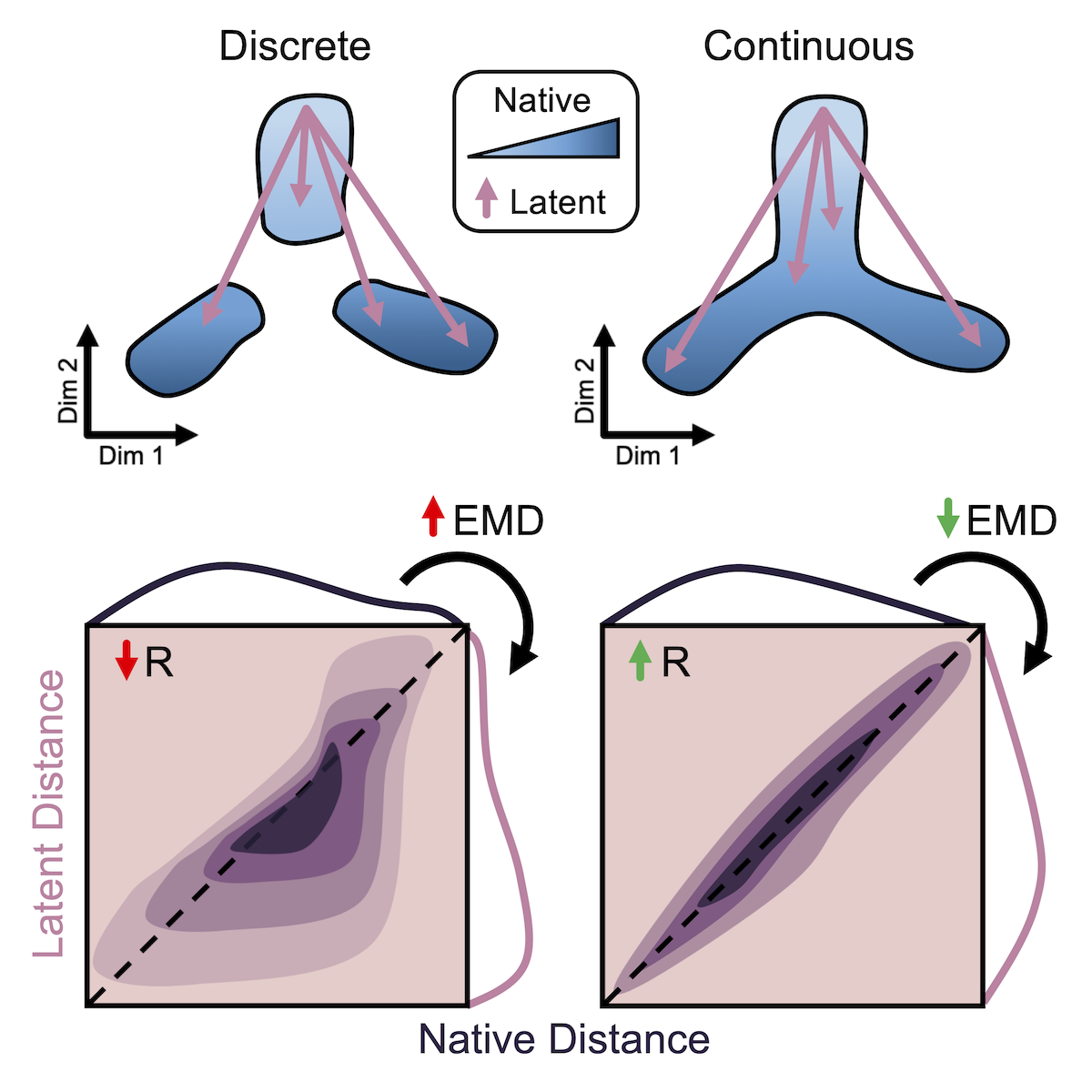
High-dimensional data is integral to modern systems biology. Computational methods for dimensionality reduction are being rapidly developed for applications to single-cell and multi-modal technologies. In order to understand what these nonlinear transformations do to the underlying biological patterns in our data, we developed a framework of quantitative metrics for global and local distance preservation. See our Cell Reports publication for details and analysis of existing dimensionality reduction methods.
Contents
fcc_utils.py
Contain utility functions for manipulating datasets and comparing feature-reduced latent spaces.
Documentation available at KenLauLab.github.io/DR-structure-preservation/.
Tutorials
Consult distance_preservation_tutorial.ipynb for info on how to perform global and local structure preservation analyses on low-dimensional embeddings of your own datasets.
Required Python Dependencies
Install using pip:
pip install -r requirements.txt
Optional Packages
In order to use the “FIt-SNE” implementation of t-SNE, you’ll need to download FFTW and compile the code from the FIt-SNE repo.
Clone the scvis and ZIFA packages and install with python setup.py install from their home directories.