
Overview
A major challenge for droplet-based single-cell sequencing technologies is distinguishing true cells from uninformative barcodes in datasets with disparate library sizes confounded by high technical noise (i.e. batch-specific ambient RNA). dropkick is a fully automated software tool for quality control and filtering of scRNA-seq data with a focus on excluding ambient barcodes and recovering real cells bordering the quality threshold.
Read our Genome Research paper for more information on the development and benchmarking of dropkick, and visit the GitHub repository to view the source code.
dropkick QC
dropkick provides a QC module for initial evaluation of global distributions that define barcode populations (real cells vs. empty droplets) and quantifies the batch-specific ambient gene profile.
You can run the dropkick.qc module from terminal for a quick look at the total UMI distribution and ambient gene profile, saved as *_qc.png:
dropkick qc path/to/counts.h5ad
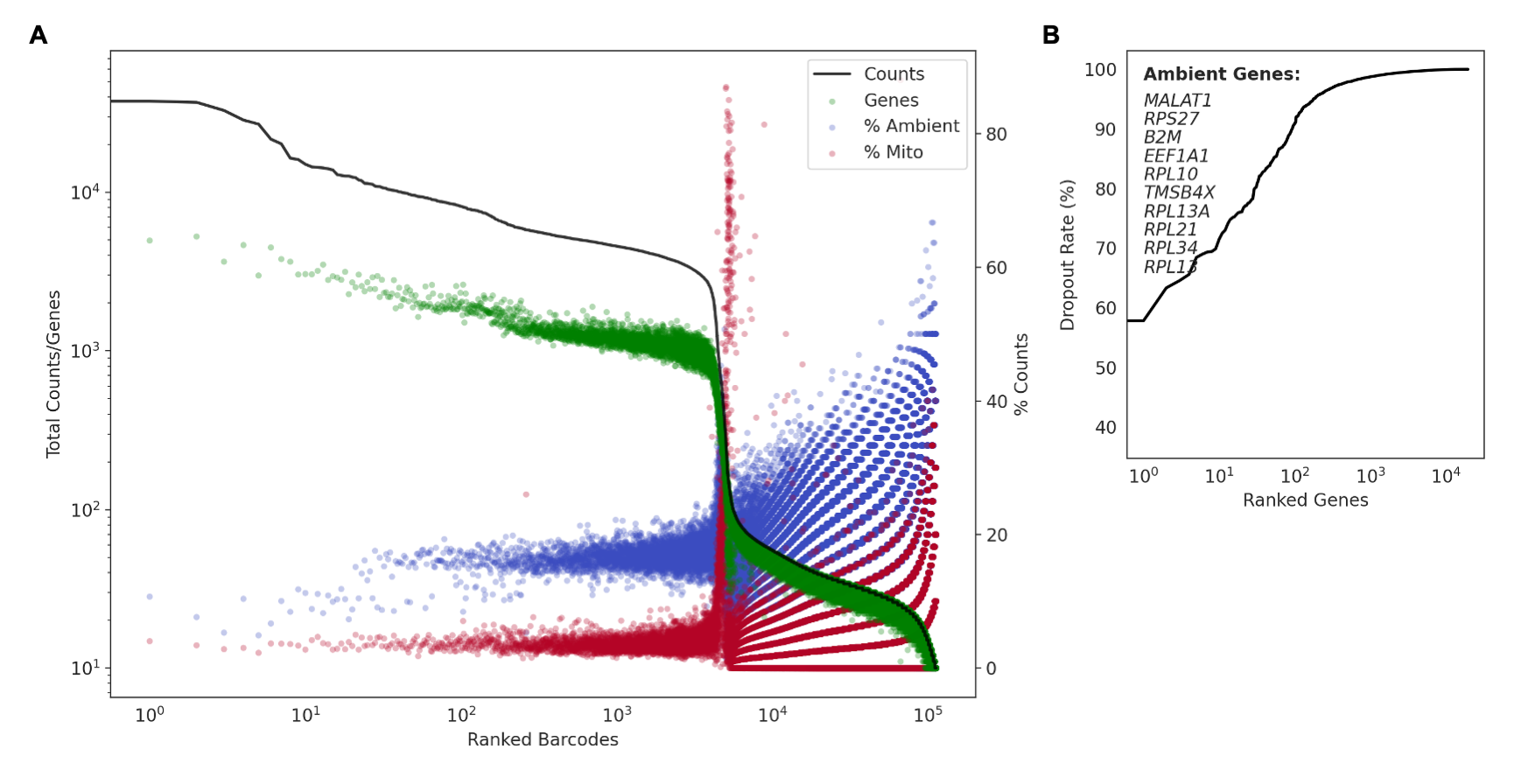
The resulting quality control report plots a log-log curve of total counts and genes per ranked barcode along with the percentage of mitochondrial and ambient counts for each barcode (A). The ambient genes are determined by ranking genes by dropout rate in ascending order and taking the top highly-expressed transcripts (B).
dropkick filtering
The primary dropkick module performs cell identification through a weakly-supervised logistic regression model. dropkick establishes initial thresholds on predictive global heuristics using an automated gradient-descent method, then trains a gene-based model to assign confidence scores to all barcodes in the dataset. dropkick model coefficients are sparse and biologically informative, identifying a minimal number of gene features associated with empty droplets and low-quality cells.
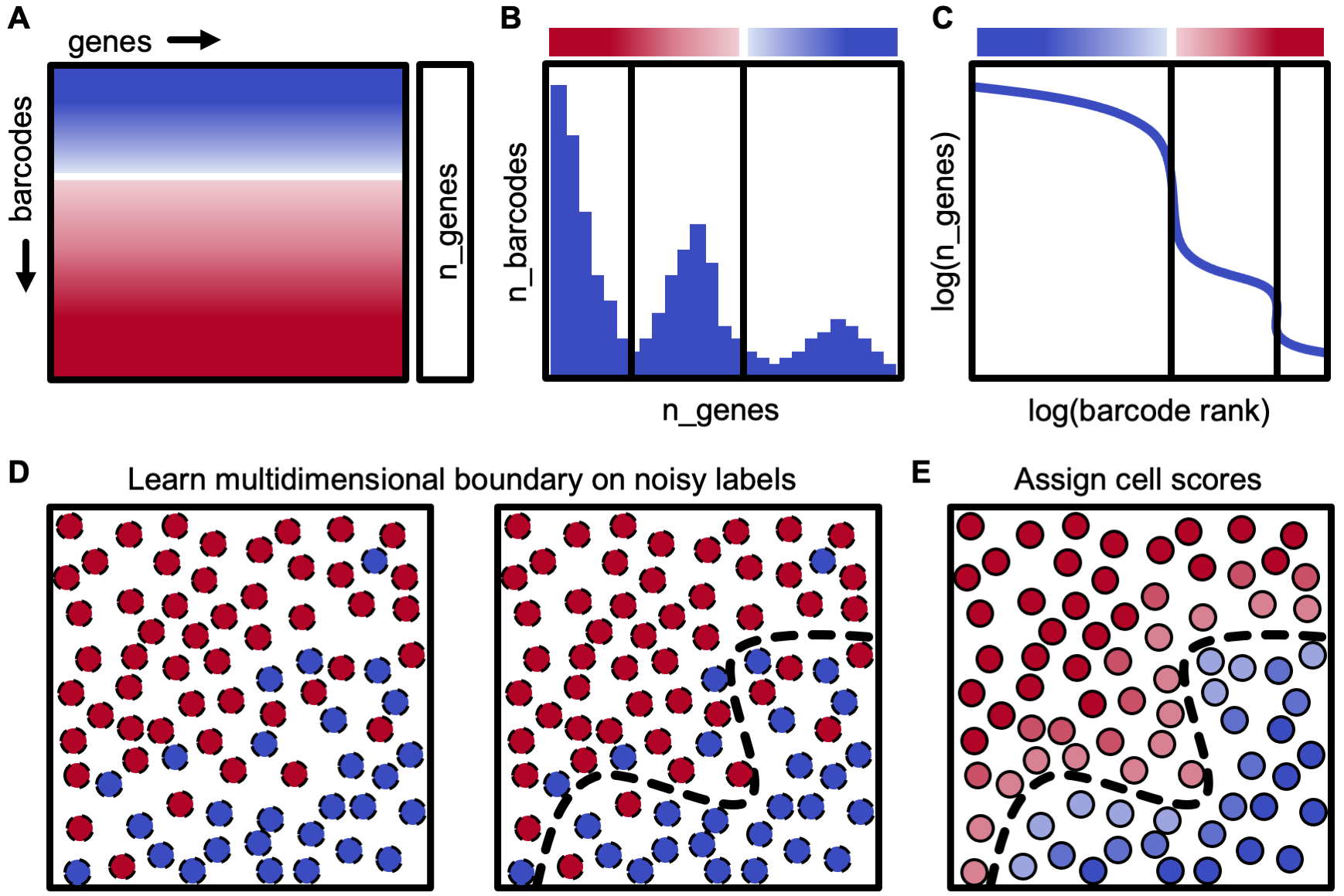
dropkick can be run as a command line tool or interactively with the scanpy single-cell analysis suite.
Usage from command line:
dropkick run path/to/counts.h5ad
Installation
Installation via pip or from source requires a Fortran compiler (brew install gcc for Mac users). The only other pre-requisite for the Python environment prior to installation is the numpy package (pip install numpy).
Install from PyPI:
pip install dropkick
Or compile from source:
git clone https://github.com/KenLauLab/dropkick.git
cd dropkick
python setup.py install
Documentation
Full documentation is available at KenLauLab.github.io/dropkick.
